13.2 Using MetaForest
To illustrate how to use MetaForest to identify relevant moderators in a small sample meta-analysis, I will apply it to the curry data.
# Load the metaforest package
library(metaforest)
# Select only the relevant variables from the curry data
data <- curry[, c("d", "vi", "study_id", "sex", "age", "location", "donorcode",
"interventioncode", "controlcode", "outcomecode")]13.2.1 Checking convergence
For any random forests model, it is important to check whether the model converges. Convergence is assessed by examining the cumulative mean squared out-of-bag prediction error (MSE), as a function of the number of trees in the model. When the MSE stabilizes, the model is said to have converged. To get an impression of how many trees are required to have the model converge, we will run the analysis once with a very high number. We then pick a smaller number of trees, at which the model is also seen to have definitely converged, to speed up the subsequent computationally heavy steps, such as replication and model tuning. We will examine convergence again for the final model.
# Because MetaForest uses the random number generator (for bootstrapping),
# we set a random seed so analyses can be replicated exactly.
set.seed(242)
# Run model with many trees to check convergence
check_conv <- MetaForest(d~.,
data = data,
study = "study_id",
whichweights = "random",
num.trees = 20000)
# Plot convergence trajectory
plot(check_conv)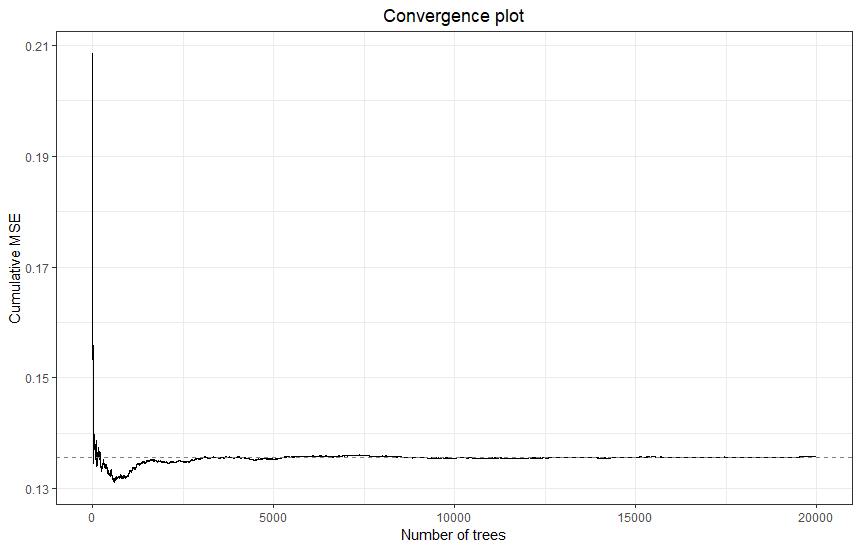
It can be seen that this model has converged within approximately 5000 trees. Thus, we will use this number of trees for subsequent analyses. We now apply recursive pre-selection using the preselect function. This algorithm helps eliminate noise moderators by running the analysis, dropping the moderator with the most negative variable importance, and then re-running the analysis until all remaining variables have positive importance. This recursive algorithm is replicated 100-fold. Using preselect_vars, we retain only those moderators for which a 50% percentile interval of the variable importance metrics does not include zero (variable importance is counted as zero when a moderator is not included in the final step of the recursive algorithm).
set.seed(55)
# Model with 10000 trees for replication
mf_rep <- MetaForest(d~.,
data = data,
study = "study_id",
whichweights = "random",
num.trees = 5000)
# Run recursive preselection, store results in object 'preselect'
preselected <- preselect(mf_rep,
replications = 100,
algorithm = "recursive")
# Plot the results
plot(preselected)
# Retain only moderators with positive variable importance in more than
# 50% of replications
retain_mods <- preselect_vars(preselected, cutoff = .5)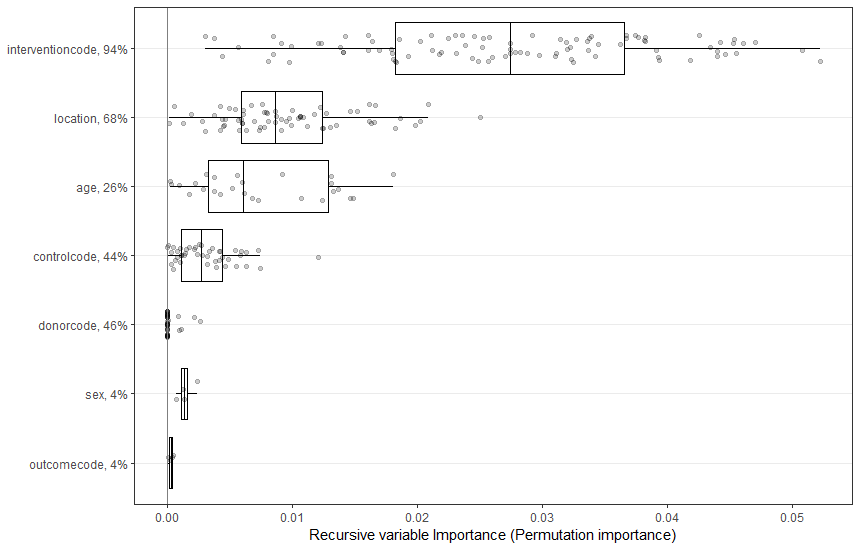
We can see that only interventioncode and location have been selected.